About

Highly skilled and versatile Full Stack Web Developer with proven expertise in designing, developing, and deploying robust applications and processes in collaboration with non-technical personnel.
Wireframes, UI, and whiteboards ideas using Figma.
Develops web applications with up-to-date frameworks and technologies. Most frequently uses NextJS + Tailwind for rapid web development.
Deploys applications to Google Cloud Platform and OpenShift/Kubernetes.
Integrates workload managers such as Slurm and Snakemake for compute-intensive tasks.
Portfolio
Primer ID
A free and open-source platform that allows researchers from all over the world to process Next Generation Sequencing data for drug resistance, sequence alignment, and outgrowth dating. This platform was created in conjunction with the pipelines found atGitHub/ViralSeq/viral_seq and GitHub/veg/ogv-dating.
This platform consists of two main infrastructure components, a web application for users to create a pipeline submission and a capable backend server for running the compute-intensive pipelines.
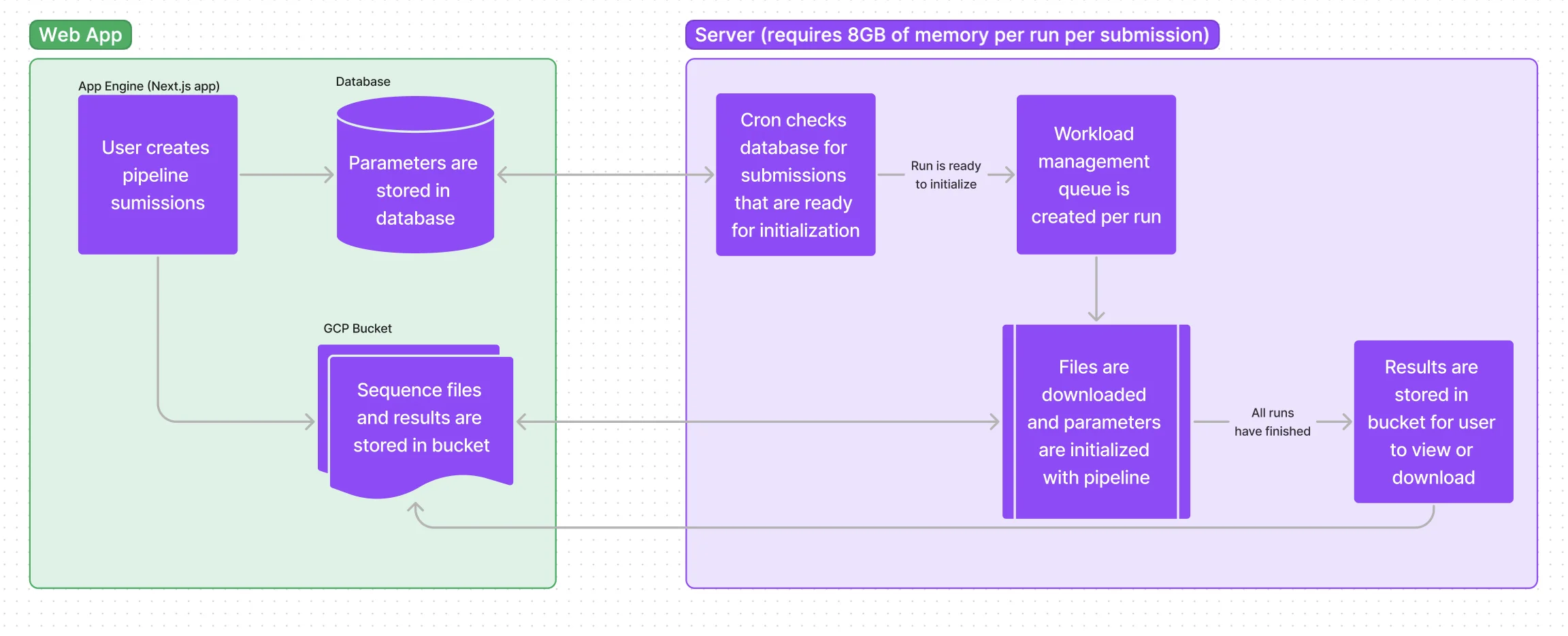
Parameters are created in the web app and stored to a database. Sequence files are uploaded to a private GCP bucket using a resumable signed url. A cron job on the cluster server pings the app API checking for submissiosn that are ready to initialize into the workload manager queue (Slurm) to await an environment in which a proper amount of memory and processing power are available to run through the pipeline. Once submissions have been processed, results are stored in the bucket and a link is emailed to the user for viewing and downloading.
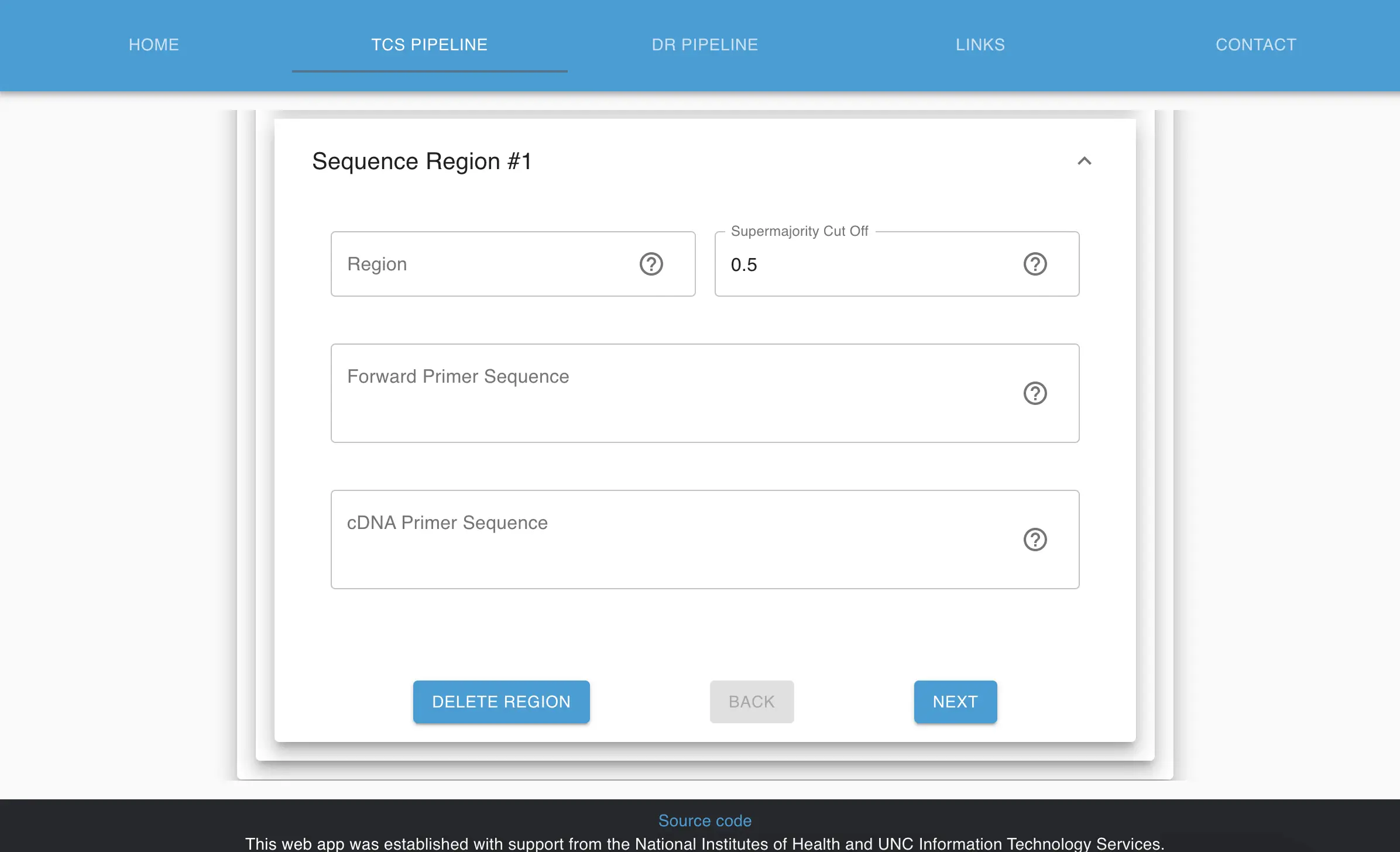
Complex parameters can be set to configure processing in the UI. I created a visually appealing way for users that have to go through this form up to 4 times or more per submission. In the research world, we very frequently use verification steps for users to check their work for precision at each step.
A Docker setup process is available to replicate this workflow on one's own computer for replication studies and continued research.
I used UNC's color palette throughout the theme and developed using Next.js + Tailwind CSS.
Phylodynamics
This platform aids in the processesing of HIV sequences from all tested cases in North Carolina. Both new and existing cases are tracked, creating a phylogeny network. The platform automates many steps of a process that were previously done manually by personell, durastically reducing turnaround times for getting results to where they are needed.
The input is a sample of a virus and this is the output we generate for every patient (no PHI in document).Server processes watch for new data to process. After processing, viewable records are created of each specimen. These records are sent to the web application where they are accessible via a searchable data table. Personnel then mark each processed specimen as successful, failed, or sent for a repeat sampling. Successful data is forwarded to a RedMine database (managed by others) in the network to be recoupled to PHI.
After the handoff of data, a team at the UNC School of Medicine pools and distributes findings. Findings include sharing outbreak hotspots with the Department of Health and Human Services so that they can ramp up public prevention as well as providing the CDC with official case numbers for the state.
*Website is access protected and repo is private.Epitope Analysis Tool
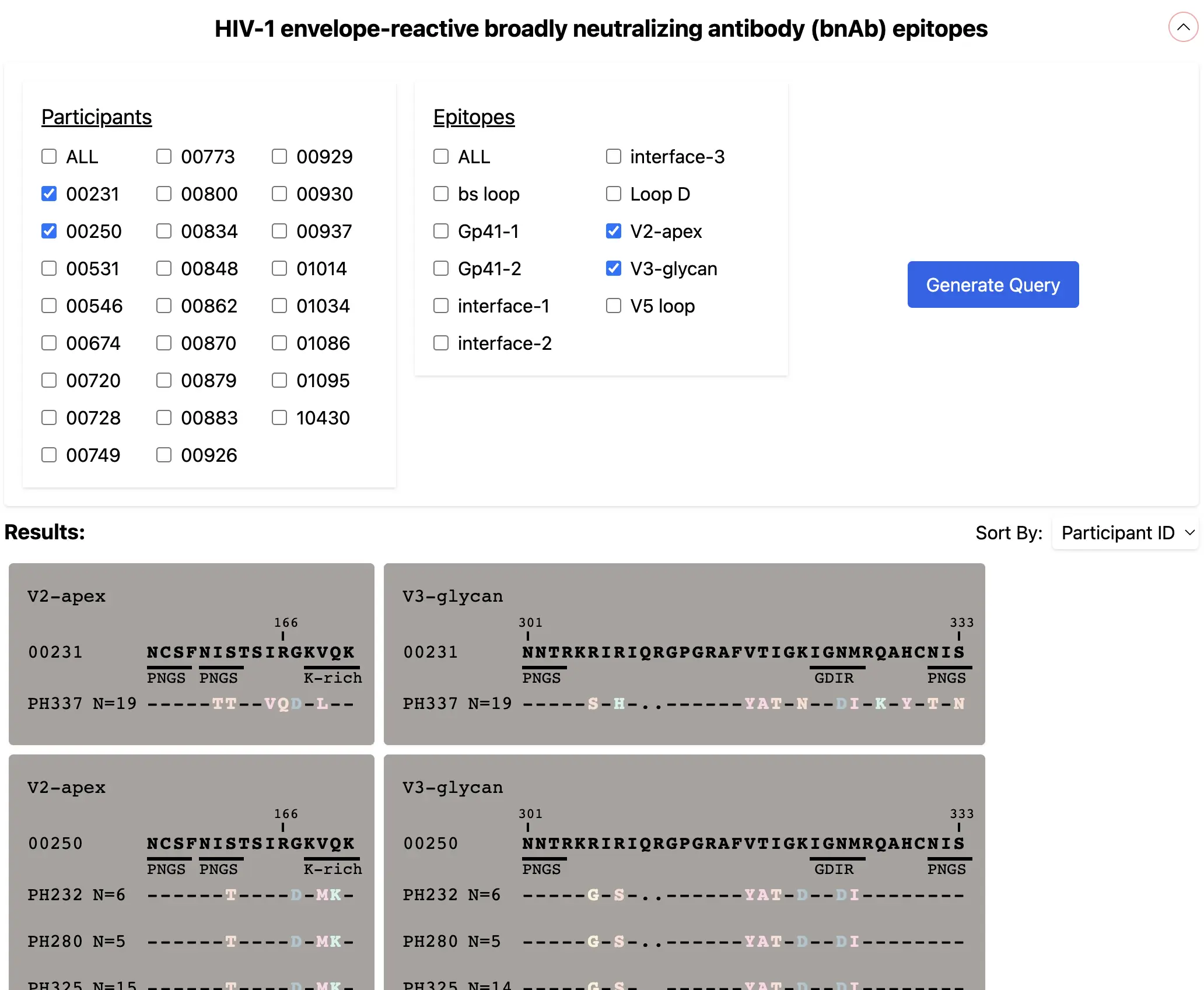
I created this prototype application to visually assist someone's grant application. An epidemiologist wants to create an interdisciplinary analysis tool between immunologists and virologists.
The whiteboard of the initial concept can be seen below. We have viral antibody information that is useful for epidemologists to visually compare. A page was needed to filter available patients and their sequences.
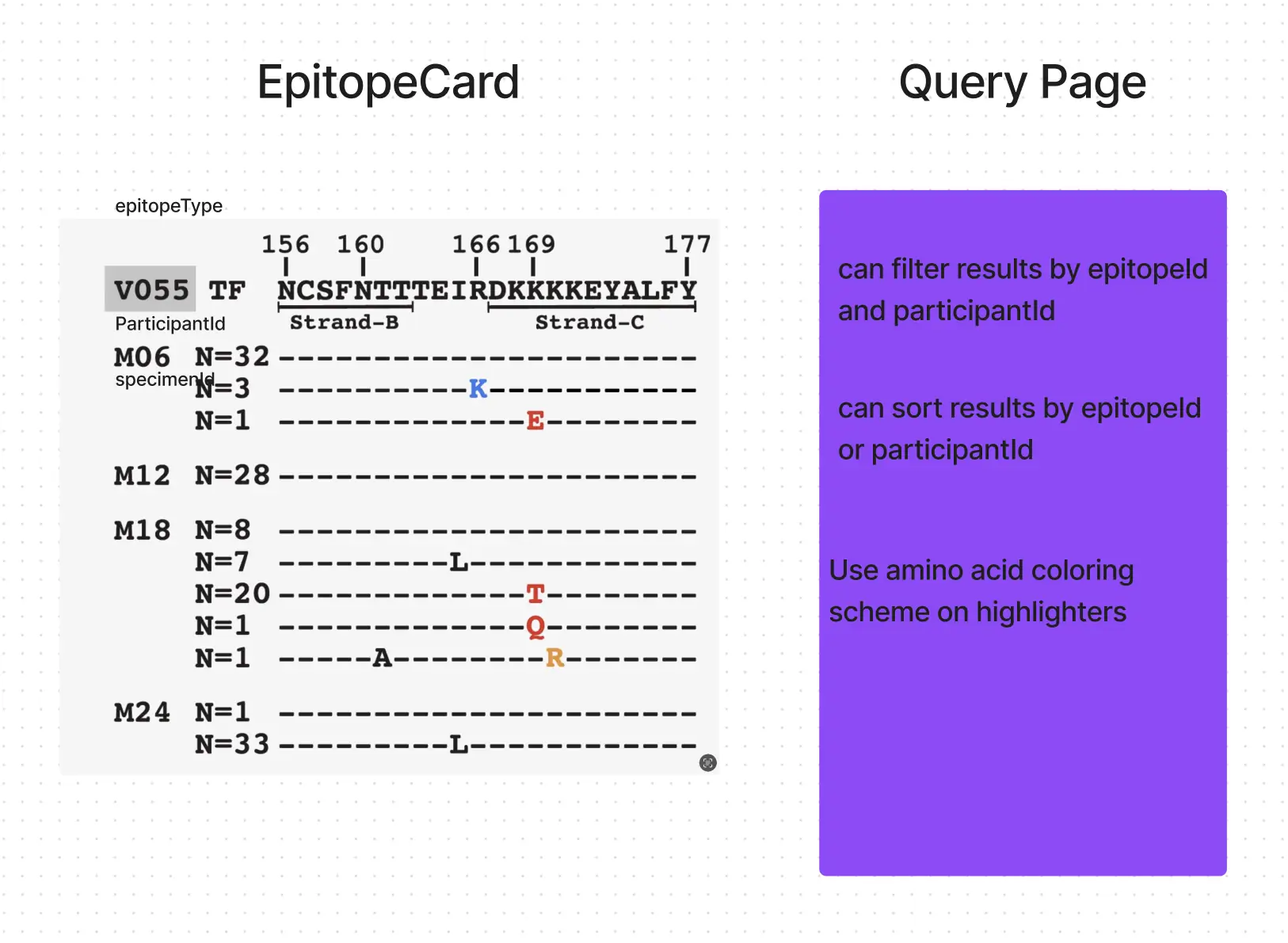
A solid structure was defined before development began:
At this early conceptual stage, many typical components of a web application can be foregone to balance cost and time for projects that may or may not get funded. Omitting a database and static hosting helped reduce cost and turnaround time for this prototype.
Cited Publications
- Zhou S, Hill CS, Clark MU, Sheahan TP, Baric R, Swanstrom R.
Primer ID Next-Generation Sequencing for the Analysis of a Broad Spectrum Antiviral Induced Transition Mutations and Errors Rates in a Coronavirus Genome. Bio Protoc. 2021 Mar 5;11(5):e3938. doi: 10.21769/BioProtoc.3938. eCollection 2021 Mar 5.
PMID: 33796612
https://pubmed.ncbi.nlm.nih.gov/33796612/ - Zhou S, Sizemore S, Moeser M, Zimmerman S, Samoff E, Mobley V, Frost S, Cressman A, Clark M, Skelly T, Kelkar H, Veluvolu U, Jones C, Eron J, Cohen M, Nelson JAE, Swanstrom R, Dennis AM.
Near Real-Time Identification of Recent Human Immunodeficiency Virus Transmissions, Transmitted Drug Resistance Mutations, and Transmission Networks by Multiplexed Primer ID-Next-Generation Sequencing in North Carolina. J Infect Dis. 2021 Mar 3;223(5):876-884. doi: 10.1093/infdis/jiaa417.
PMID: 32663847
https://pubmed.ncbi.nlm.nih.gov/32663847/ - Shuntai Zhou, Collin S. Hill, Ean Spielvogel, Michael U. Clark, Michael G. Hudgens, Ronald Swanstrom
Unique Molecular Identifiers and Multiplexing Amplicons Maximize the Utility of Deep Sequencing To Critically Assess Population Diversity in RNA Viruses. ACS Infectious Diseases Article ASAP. DOI: 10.1021/acsinfecdis.2c00319
PMID: 36326446
https://pubmed.ncbi.nlm.nih.gov/36326446/ - Shuntai Zhou, Nathan Long, Matt Moeser, Collin S Hill, Erika Samoff, Victoria Mobley, Simon Frost, Cara Bayer, Elizabeth Kelly, Annalea Greifinger, Scott Shone, William Glover, Michael Clark, Joseph Eron, Myron Cohen, Ronald Swanstrom, Ann M Dennis
Use of Next Generation Sequencing in a State-Wide Strategy of HIV-1 Surveillance: Impact of the SARS-CoV-2 Pandemic on HIV-1 Diagnosis and Transmission J Infect Dis. 2023 Jun 7 DOI: 10.1093/infdis/jiad211
PMID: 37283544
https://pubmed.ncbi.nlm.nih.gov/37283544/
Effects + Data Visualization
Creating visual effects helps me retain my foundational math skills. I have a Three.js + Fiber Portfolio as well as an account at Observable where I have outlined a few animations.
My latest example of data visualization is adding an output file to a sequencing pipeline where I use HTML + Google Charts to display the output in graphs and charts.
Viral sequences displayed as drug restistance mutations and their recency.notesy.app
I set out on a mission to fine-tune my UI-design and Figma skills. Over just a few weeks I created a note taking app, borrowing the best ideas from current writing platform designs and features.
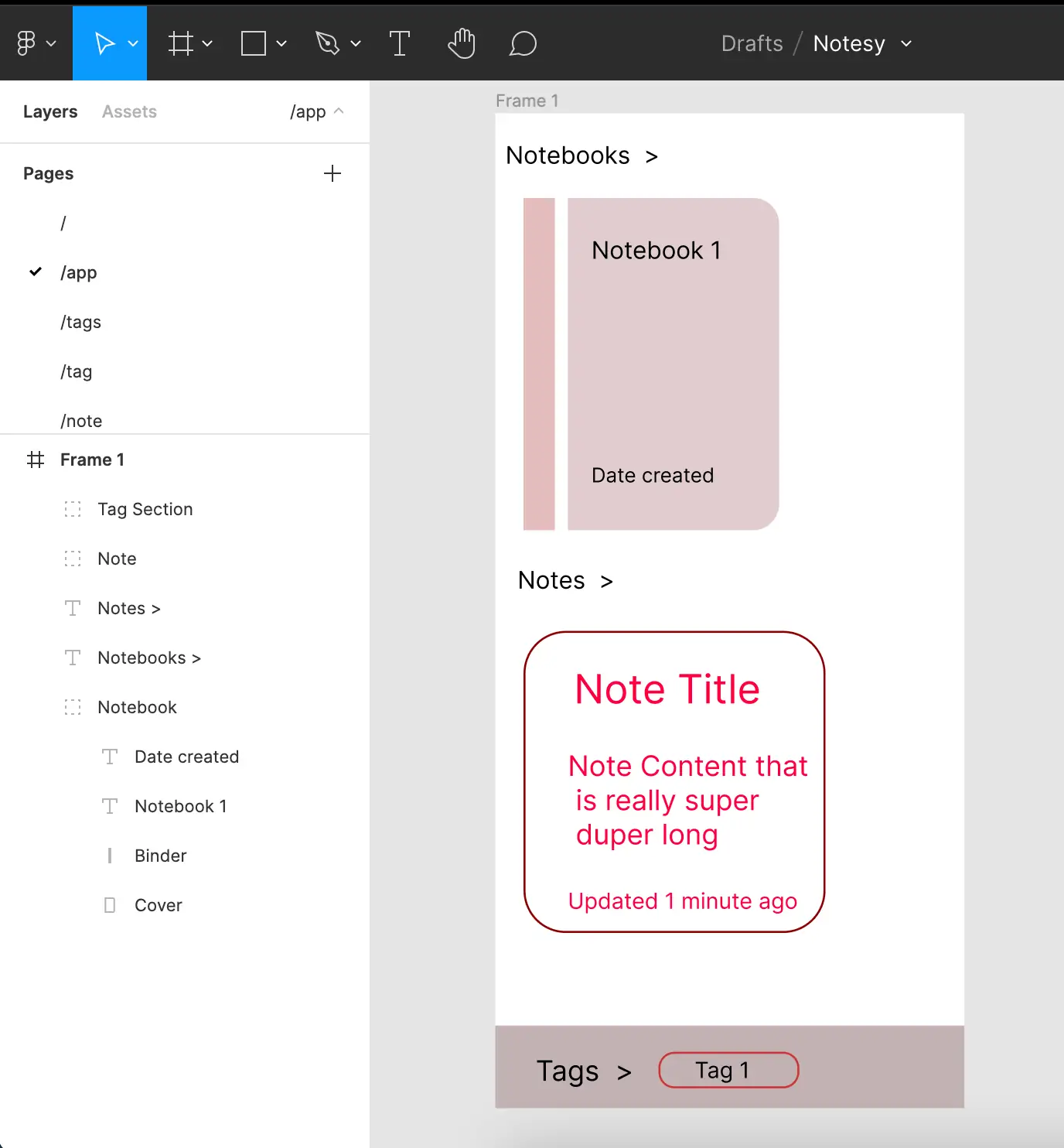
I focused on a simple, easy-to-use interface. I created a marketing page which outlines the application and its features. The onboarding process is smooth and simple. I jam-packed the FAQ page with SEO-rich content.
- Lighthouse scores considered (performance at 100)
- Customized DraftJS saved to MongoDB to retain notes
- Websockets for shared editing
- React useReducer and custom hooks
- List virtualization
- NextAuth authentication
- Installable as a Progressive Web App
App can be found at notesy.app

react-zoom-scroll-effect
I noticed this behavior out in the wild and found that there were no packages or css utilities that take advantage of scrolling. So I took 3 days to make this really fun React component into an NPM package.
There are a few variations, parameters, and uses for this visual feature so this was a prime candidate for StoryBook testing.
I used Rollout to minify and package, as it appeared to be the tried-and-true option for NPM deployment.
The package and documentation can be found here.
Sheet Music Creator
A platform to create a custom format of sheet music intended for students with a focus on learning disabled students. I freelanced with a non-technical team that wanted to expand their efforts in creating this sheet music and sought to automate and digitize the process.
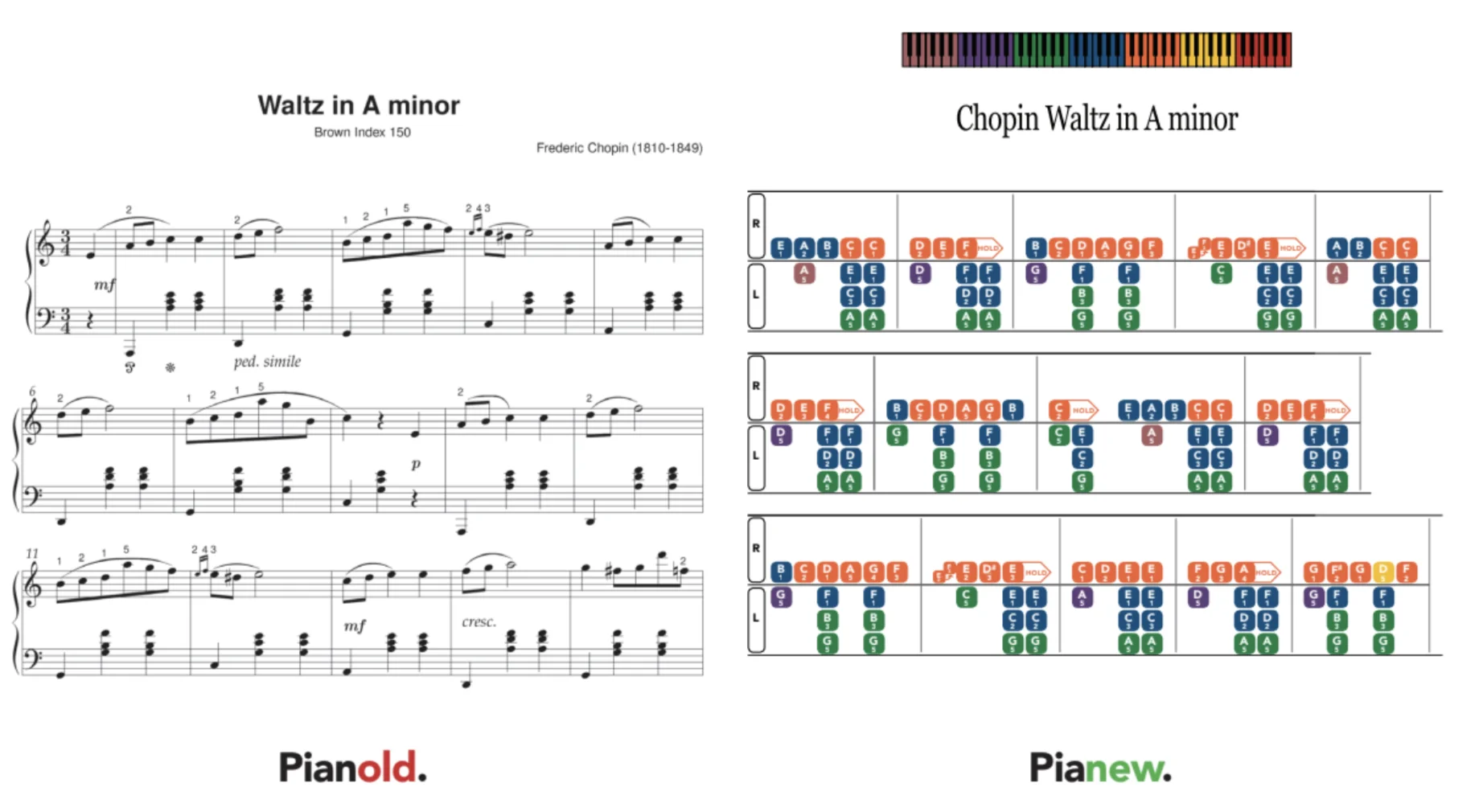
The main technical requirement from the client was that it had to be installable on the workers laptops. Along with the other specifications of this project, it turned out to be a great fit for a Progressive Web App.
Main feature of application: a drag-and-drop interface for creating the custom sheet music.Development consisted of a React frontend with heavy usage of the NPM drag-and-drop package react-dnd to edit tabs. Tab objects stored in a MongoDB collection. Completed tabs were converted to PDF and saved to a storage bucket for user viewing.
My background with music helped to hit the ground running with this team. In my adult years I attended two years at MuziekSchool Roeselare, a music school in Belgium for students of all ages, where I learned to converse about music in English, Dutch, and French.
*Website is access protected and repo is private.CaaS
Many of my projects are containerized and documented for easy research replication. This also makes it easier to manage deployment into a Kubernetes cluster. I usually start with a template as diagrammed below.
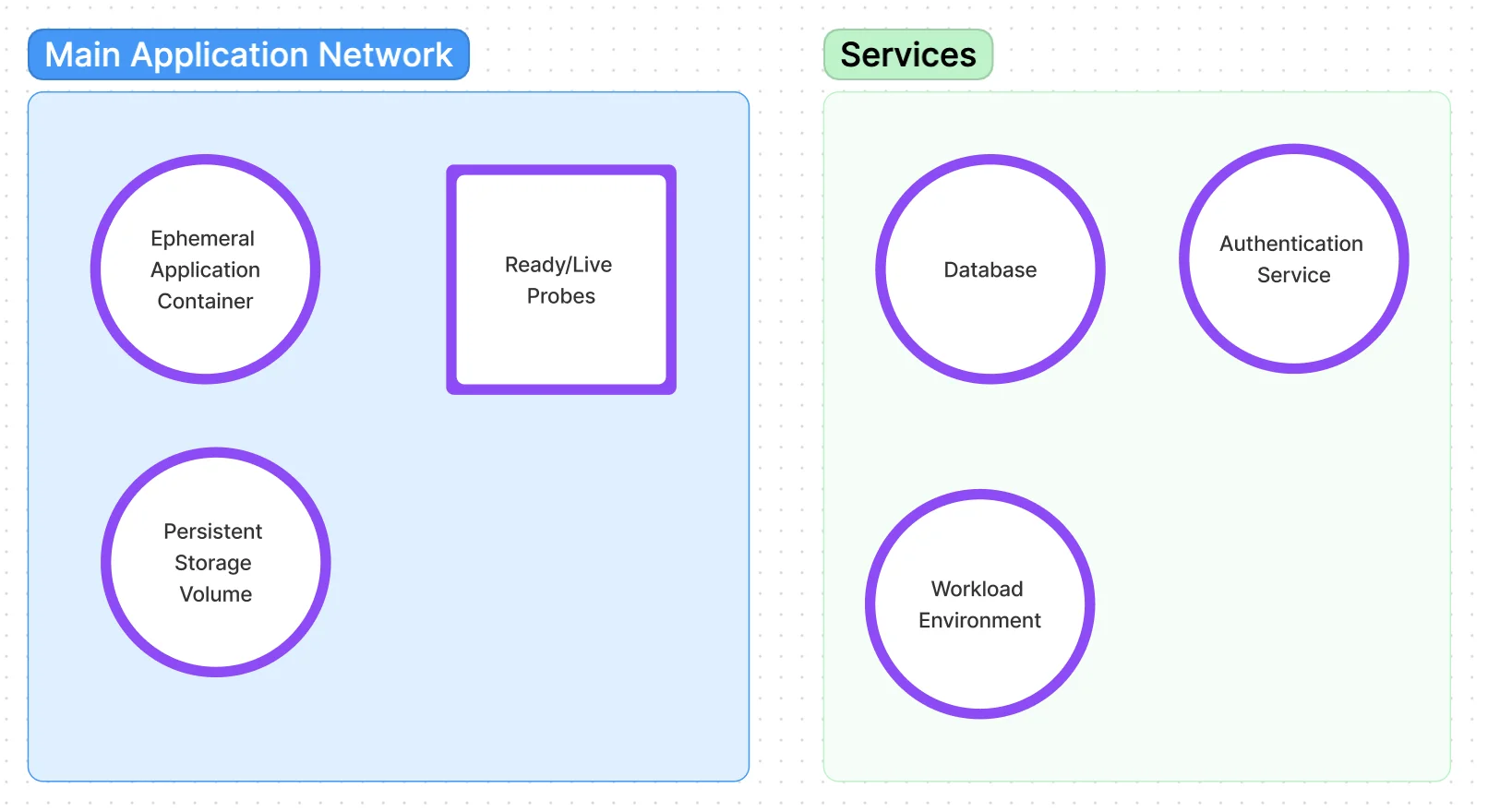
I started using OpenShift in version 1 before there was a higher level developer view so I learned all of the administrator details related to building an image, deploying a pod, and registering readiness probes and health checks.
And what kind of developer would I be if I didn't automate backups of all my databases to a storage volume.
High Performance Computing
Working with researchers means cutting edge ideas requiring intense computational power.
In one process that I have automated, users can upload 20+ files per batch with each file being measured in gigabytes. Each file needs a list of memory-intensive string operations performed on it. Running these processes in sequence would further delay receiving results. After ensuring these computations are parallelable, I queue them up with a workload manager to execute.
These operations would clog up a normal server depending on its performance capabilities. For less intense computations, this would be ideal for serverless nodes (AWS lambdas).
Server usage requires crystal clear communication with IT department to retain security regarding permissions, installations, and data I/O.
Cloud
Many of the latest web & bioinformatics cloud tools would be applicable to my projects needs, though UNC has a comparable IT hardware infrastructure. Keeping a budget in mind, we prefer on-campus hardware until projects reach a certain threshold of computation and usage.
My university is best integrated with Google Cloud Platform. I participated in a work event where the IT department went to Google's Chapel Hill headquarters to receive five days of training - we learned procedures like spinning up a compute engine and using signed URL's to read/write in a private bucket, tasks that I still frequently use.